1st IEEE Symposium on Biological Data Visualization 2011 and
BMC Bioinformatics
 | HiTSEE: A Visualization Tool for Hit Selection and Analysis in High-Throughput Screening Experiments |
Enrico Bertini
Hendrik Strobelt
Joachim Braun
Oliver Deussen
Ulrich Groth
Thomas U. Mayer
Dorit Merhof
University of Konstanz, Germany
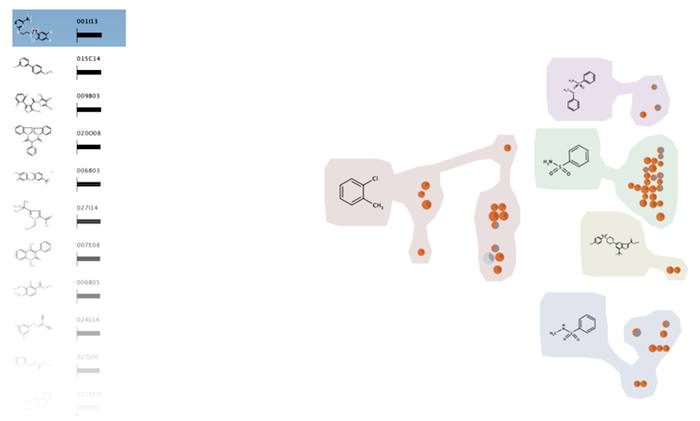
Abstract
We present HiTSEE (High-Throughput Screening Exploration Environment) a visualization tool for the analysis of large chemical screens for the analysis of biochemical processes. The tool supports the analysis of structure-activity relationships (SAR analysis) and, through a flexible interaction mechanisms, the navigation of large chemical spaces. Our approach based on the projection of one or few molecules of interest and the expansion around their neighborhood allows for the exploration of large chemical libraries without the need to create an all encompassing overview of the whole library. We describe the requirements we collected during our collaboration with biologists and chemists, the design rationale behind the tool, and two case studies.
Citation
Please use this citation:@Article{22607449,
AUTHOR = {Strobelt, Hendrik and Bertini, Enrico and Braun, Joachim and Deussen, Oliver and Groth, Ulrich and Mayer, Thomas and Merhof, Dorit},
TITLE = {HiTSEE KNIME: a visualization tool for hit selection and analysis in high-throughput screening experiments for the KNIME platform},
JOURNAL = {BMC Bioinformatics},
VOLUME = {13},
YEAR = {2012},
NUMBER = {Suppl 8},
PAGES = {S4},
URL = {http://www.biomedcentral.com/1471-2105/13/S8/S4},
DOI = {10.1186/1471-2105-13-S8-S4},
PubMedID = {22607449},
ISSN = {1471-2105},
ABSTRACT = {We present HiTSEE (High-Throughput Screening Exploration Environment), a visualization tool for the analysis of large chemical screens used to examine biochemical processes. The tool supports the investigation of structure-activity relationships (SAR analysis) and, through a flexible interaction mechanism, the navigation of large chemical spaces. Our approach is based on the projection of one or a few molecules of interest and the expansion around their neighborhood and allows for the exploration of large chemical libraries without the need to create an all encompassing overview of the whole library. We describe the requirements we collected during our collaboration with biologists and chemists, the design rationale behind the tool, and two case studies on different datasets. The described integration (HiTSEE KNIME) into the KNIME platform allows additional flexibility in adopting our approach to a wide range of different biochemical problems and enables other research groups to use HiTSEE.},
}Video
Downloads

|
Paper BMC Article (open source) |